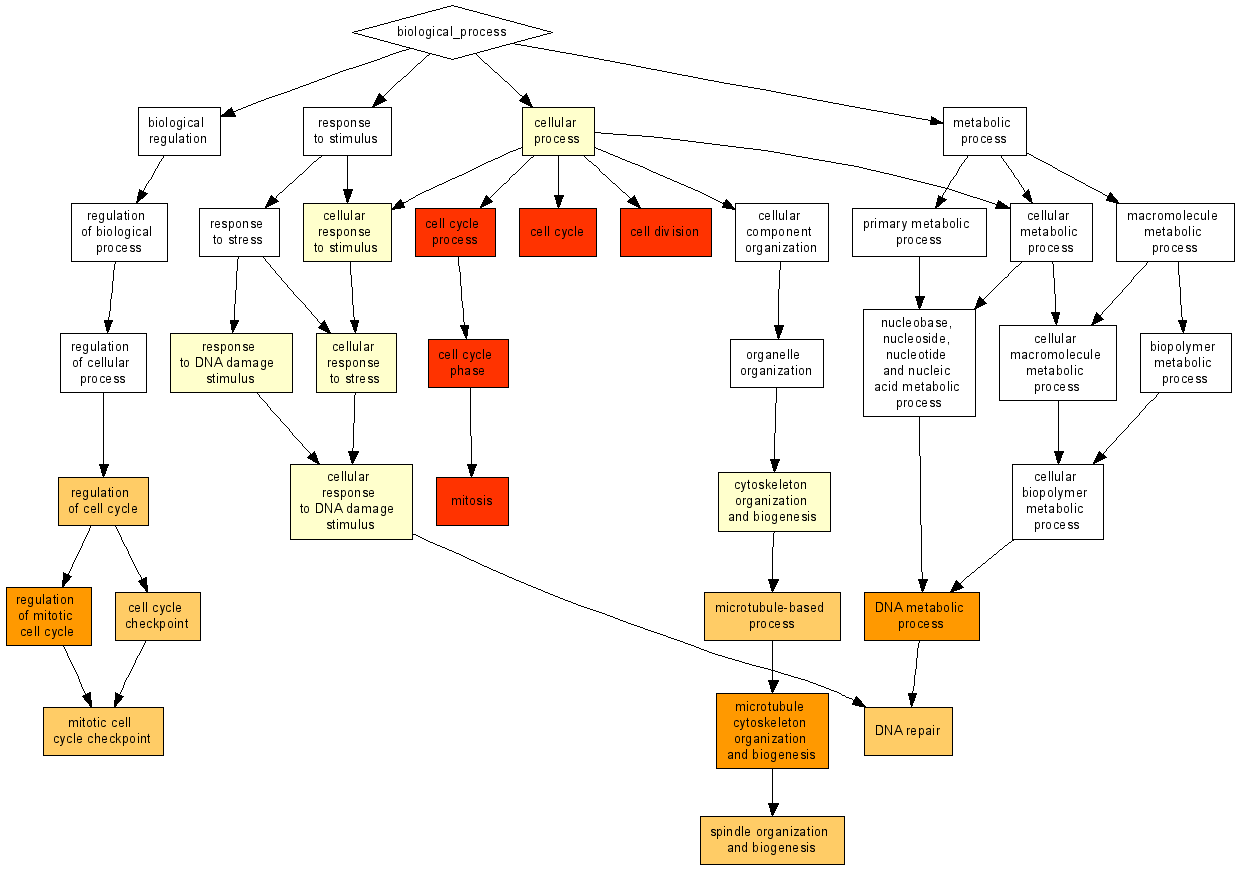
Running example
Running GOrilla is easy. Just follow the example below:
Step 1: Choose "Homo sapiens"
Step 2: Choose "Single ranked list of genes"
Step 3: Paste the contents of this file to the textbox.
This example is the list
of differential expressed genes between patients with less than 5 years to metastases and at least five years disease-free.
The data is from the van‘t Veer paper and can be
downloaded from here .
Step 4: Choose the gene ontology category - "Process"
You can adjust the sensitivity of results using the "P-value threshold". Change threshold to 10^-5.
Click on "Search enriched GO terms"
That's it. The following figure should appear on the screen: