
 |
GORILLA |  |
| Gene Ontology enRIchment anaLysis and visuaLizAtion tool | ||
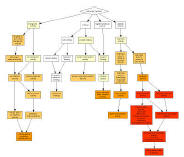
GOrilla is a tool for identifying and visualizing enriched GO terms in
ranked lists of genes.
It can be run in one of two modes:
Running example Usage instructions GOrilla News References Contact
If you choose to use this application please cite:
Graphics are generated using Graphviz.
Components of the GOrilla system were designed as part of the European Union FP6 funded Multi Knowledge Project.

Creating New Knowledge In Networks of Medical Research
Please send your comments, suggestions and bug reports to Roy Navon at: rnavon@gmail.com and include a link to your results page.