|
miTEA
miRNA Target Enrichment Analysis
|
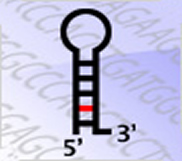 |
miTEA is a tool for identifying and visualizing enriched miRNA targets in ranked lists of genes.
It searches for enriched miRNA targets that appear densely at the top of a ranked list of genes.
For further details see
References.
Running
example
Usage instructions
References
References
If you choose to use this application please cite:
- Israel Steinfeld, Roy Navon, Robert Ach, Zohar Yakhini. "miRNA target enrichment analysis reveals directly active
miRNAs in health and disease",
Nucleic Acids Research 2012.
- Eran Eden*, Roy Navon*, Israel Steinfeld, Doron Lipson and Zohar Yakhini. "GOrilla: A
Tool For Discovery And Visualization of Enriched GO Terms in Ranked Gene
Lists",
BMC Bioinformatics 2009, 10:48.
Further reading:
- Eran Eden, Doron Lipson, Sivan Yogev, Zohar Yakhini. "Discovering
Motifs in Ranked Lists of DNA sequences",
PLoS Computational Biology, 3(3):e39, 2007.
- Enerly E*, Steinfeld I*, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA,
Johnsen H, Navon R, Rødland E, Mäkelä R, Naume B,
Perälä M, Kallioniemi O, Kristensen VN, Yakhini Z,
Børresen-Dale AL. "miRNA-mRNA Integrated Analysis Reveals Roles for
miRNAs in Primary Breast Tumors.",
PLoS One. 2011 Feb 22;6(2):e16915.
Graphics are generated using
Graphviz.
Please send your comments, suggestions and bug reports to Roy Navon at:
For questions regarding your results, please include a link to your results page.